

David M. Gilbert, Florida State University
Raymond Gosling/King’s College London
Richard Wheeler, CC BY-SA
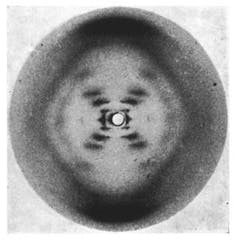
For biologists everywhere, April 25 is auspicious. It is DNA Day and commemorates the date in 1953 when scientists Francis Crick, Rosalind Franklin, James Watson and Maurice Wilkins published seminal scientific papers describing the helical structure of the DNA molecule. In 2003, April 25 was used to announce the completion of the Human Genome Project. Now annual festivities on this day celebrate the molecule of life with new discoveries. What better time to provide a new picture of DNA.
I am DNA DAVE (or at least my license plate since 1984 says so), and one of the things my lab likes to do is to “see” DNA. We take images of DNA so that we can directly measure things that are difficult to quantify using indirect methods that usually involve sequencing the four chemical units of DNA, called bases.
For example, I would like to know where on each chromosome the process of DNA replication begins. Error-free duplication of DNA is essential for producing healthy cells. When this process is incomplete or disrupted, the result can cause cancer and other diseases.
In our image that familiar double helix staircase is not visible because this perspective is zoomed out – like looking at the map of a country versus a city. Also each of these molecules is equivalent to 50,000 turns of the helical staircase – a substantial segment of a human chromosome.
Making a map of DNA
David Gilbert/Kyle Klein, CC BY-ND
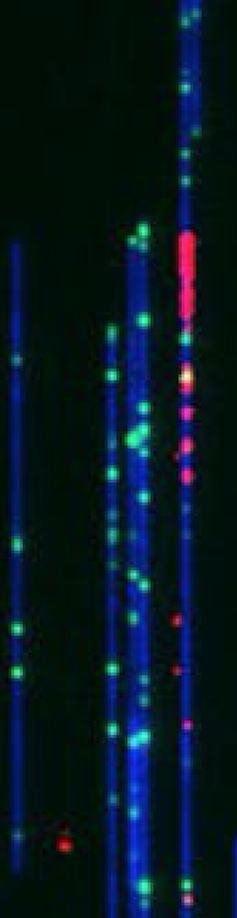
This image, taken with a device called the Bionano Genomics Saphyr imager, features individual DNA molecules – colored in blue, green and red. These strands of DNA have been aligned by threading them through narrow tubes – called nanochannels – that fit only one piece of DNA. As the DNA slips into the tube, the strands straighten.
The whole DNA molecule is colored blue and the green tick marks are landmarks – or specific sequences of DNA that occur on average every 4,500 base pairs. The pattern of landmarks provide a unique fingerprint which tells us where we are along the length of a chromosome. The red fluorescent blips tag the locations where the DNA has begun to replicate. These sites are called “origins of replication” and are where the DNA first unwinds so that the duplication process can start.
Researchers at Bionano Genomics in San Diego developed this nanochannel technology to chart regions of chromosomes that were otherwise unmappable, due to tricky genetic sequences that make it difficult to determine the order of the four bases. This device solved the problem by “looking” at the arrangement of sequences on one molecule at a time and is able to read 30 billion base pairs in one hour – the equivalent of 10 human genomes.
My team and that of Nick Rhind at the University of Massachusetts recognized that this nanochannel technology would allow us to conduct an experiment never attempted before: map all the locations where DNA replication begins simultaneously on millions of single DNA fibers.
Before a cell can divide into two independent cells, the DNA must make a copy of itself so that each one receives a complete set of chromosomes. To understand how the genetic material is duplicated it is essential to know where along the chromosome the process begins. That has been the greatest challenge to studying how the replication of our own chromosomes takes place and consequently what is going wrong in so many diseases, like cancer, in which replication goes awry.
DNA replication and cancer
Soleil Nordic/Shutterstock.com
Origins of replication have been elusive because they occur at many sites on different molecules so we need to look at single DNA molecules to detect them. Although scientists have been able to see single DNA molecules since the early 1960s, we could not tell where in the chromosomes any molecule came from so we could not map anything.
Kyle Klein, a Ph.D. student in my lab, labeled living human stem cells with red fluorescent molecules that marked locations where DNA replication was taking place, which were mapped with the Bionano device. These images were then superimposed onto the blue and green DNA maps of the same DNA molecules.
We expect this method to completely transform our understanding of how human chromosomes replicate. Moreover, since most chemotherapy drugs for cancer treatment and most carcinogens – or cancer-causing chemicals – in our environment work by attacking DNA when it replicates, we expect this method to provide a rapid and comprehensive test for how these chemicals disrupt DNA replication. We also hope it reveals how we might alleviate these negative consequences, and how we might develop better and less toxic chemotherapy treatments.![]()
David M. Gilbert, Professor of Molecular Biology, Florida State University
This article is republished from The Conversation under a Creative Commons license. Read the original article.



















